We held our first meeting with the Advisory Board (AB) on Apr 23, 2018, via a conference call. All four project team and six AB members were in attendance. The meeting was one of the milestones for the project’s first year in full operation, which ended in April. The expertise and research interests of the members of our AB span a wide spectrum across the biological and computational sciences:
- Jim Balhoff (RENCI - Renaisssance Computing Institute), ontology developer and computational biologist
- David Baum (University of Wisconsin, Madison), botanist and systemtic biologist
- Holly Bik (University of California, Riverside), microbiologist, nematologist, and genomics-based biodiversity researcher
- Chris Mungall (Lawrence Berkeley National Laboratory), ontology develper and computatioanl biologist
- Susan Perkins (American Museum of Natural History), parasitologist, systematic biologist
- Michael Sanderson (University of Arizona), botanist and systematic biologist
The main goal of the meeting was to introduce the AB to our project, including its major motivations, deliverables, and challenges, and to highlight where we’re currently at, as well as what we’re planning to do over the coming 12-18 months. Our other goal was to elicit feedback that would indicate where we might be missing opportunities, heading in a wrong direction, and where how we name and communicate what we do might cause confusion.
After summarizing the theoretical backgroun and major motivations of our project, we walked the AB through three major use cases (which are at this point “only” aspirational):
- querying GBIF using phyloreferences on the Open Tree of Life (OTL),
- quantifying the resolvability of phyloreferences (on OTL), and
- comparing relationships among phyloreferences on two phylogenies.
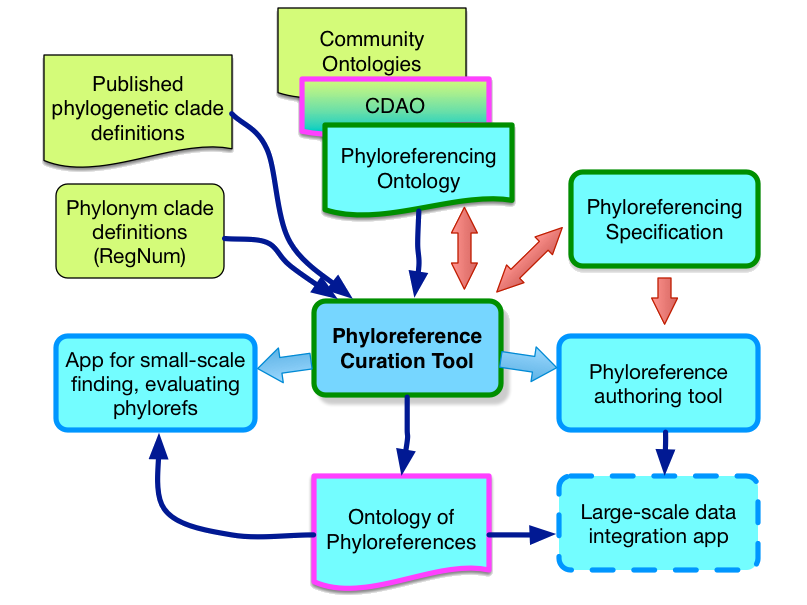
In the remaining part, we explained roadmap and deliverables, followed by demonstrating, as our first significant software product nearing its initial release, the curation tool. Going forward, we explained what we perceive as the biggest challenges facing us, namely (1) an algorithmic method for matching specififiers and dealing with missing specifiers with sufficient sophistication, (2) use case and technology development for a large-scale data integration application, and (3) engagement with the clade-oriented trait evolution and trait biodiversity research community.
Overall the meeting was very successful and productive, as measured by the feedback and thoughts that the AB volunteered. Members of the AB asked questions throughout, including high-level, big picture questions, and shared greatly valuable suggestions afterwards in email. The feedback gave us a lot to think about and to consider how we can best act on the ideas, including particular opportunities for engaging the authors of published clade definitions with our work, and opportunities for better synergizing with the RegNum database. One piece of feedback has already led to action. We decided to clarify how we name and thus refer to one of our next major product milestones: what we used to call the Ontology of Phyloreferences (prompting confusion with the Phyloreferencing Ontology, occasionally even amongst ourselves) is now the Ontology of Phylogenetic Clade Definitions (because that’s what it really is meant to be), or the Clade Ontology in short (leading to CLADEONT as an appealing candidate for the identifier prefix for terms in the ontology).
We’re now looking forward to our next meeting with the AB, which we plan to hold in about a year (i.e., about one year prior to the anticipated end of funding from the current grant) as an in-person face-to-face meeting, to facilitate more in-depth discussion and brainstorming.