About phyloreferencing
Phyloreferencing aims to provide a standard mechanism for defining biological clades with precise and fully machine-processible semantics. The name is in analogy to the ubiquitous mechanism of georeferencing, which instead of ambiguous geographic place names uses geographic coordinates to enable powerful computation for geography-linked data.
Phyloreferencing uses machine-interpretable definitions of the unique pattern of evolutionary descent that distinguishes a clade from all others. A clade defined in this way can include thousands of species, or can identify a monophyletic subgroup within a species. In contrast to authoritative nomenclatural naming, our goal is to allow users to construct phyloreferences instantly for any group of shared evolutionary descent for which they wish to communicate (or obtain) data.
Why phyloreferencing
Traditionally, scientists have used taxon names based in Linnaean nomenclature to communicate, obtain, and aggregate organism-linked data. However, doing so suffers from serious limitations. The semantics of suchtaxon names are too often ambiguous, subject to divergent interpretation, and, perhaps most importantly, unavailable to computation. And with the advent of next-generation sequencing, metagenomics, and other modern biological data collection technologies, many groups of organisms for which we have interesting data do not yet, and may never have a Linnaean name. Hence, Linnaean names fundamentally hamper our ability to communicate and aggregate organism-linked data freely, and precisely.
Phyloreferencing overcomes these limitations by defining ontology-based references to elements on the Tree of Life that are unambiguous and the semantics of which are amenable to powerful machine reasoning, making clade definitions fully computable.
A full account of the rationale and arguments for phyloreferencing can be found in Cellinese et al. (2022) “Phyloreferences: Tree-Native, Reproducible, and Machine-Interpretable Taxon Concepts”, Philosophy, Theory, and Practice in Biology 14:7.
History
The roots of Phyloreferencing as an idea go back to a group of scientists at the 2009 Phyloinformatics VoCamp (a codefest-type event for vocabulary and ontology development), held in conjunction with TDWG 2009, the annual conference of the Biodiversity Information Standards organization. Leaning on the PhyloCode for symbolism, the group, which included Nico and Hilmar, developed ways to express clade definitions as metadata queries against a database of phylogenetic trees.
The core informatics ideas underlying this project, namely using formal ontologies and machine reasoning to express universally computable clade definitions, go back in large part to the RegNum database of phylogenetic clade definitions, and the work of Phenoscape on using formal ontologies and machine reasoning to enable large-scale computing with phenotype observations across disparate sources and communities of practice. The fundamental issues in using names to communicate data about anatomical concepts (such as phenotypes) turn out to be very similar to those faced in using (Linnaean) names to communicate data about groups of organisms. In a way, we translate the techniques developed for computing with phenotype descriptions to revolutionize the ways we can use computation to integrate data by groups of organisms, unconstrained by the limitations of nomenclature.
As its theoretical underpinnings, our work builds on more than a decade’s worth of theoretical and applied research into phylogenetic taxonomy.
Logo
The Phyloreferencing logo artwork was designed by Kim Schoonover and made possible by a gift from Ken and Linda McGurn. It is available for use under a CC-BY 4.0 license (as is the rest of the content here).1 Other formats than the ones shown below are available.



Acknowledgements
Image credits
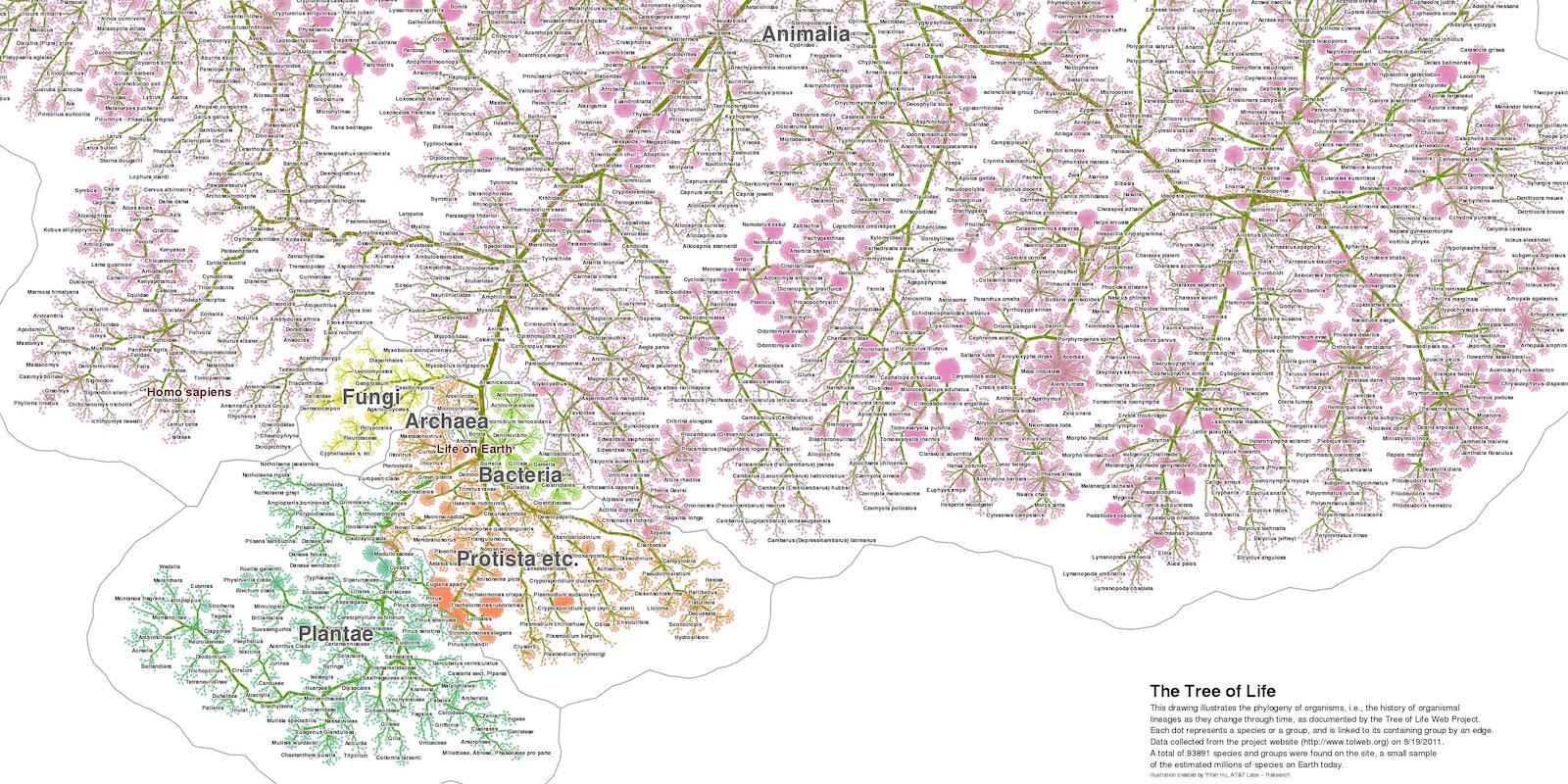
The Tree of Life visualization, which also forms the backdrop on the front page and serves as standard image for blog posts, is reused with permission from Yifan Hu. The visualization uses Web Tree of Life data. You can read more about it and find the original here: http://yifanhu.net/TOL/.
Funding

This project is funded by the US National Science Foundation through collaborative grants DBI-1458484 and DBI-1458604 to Hilmar Lapp (Duke University) and Nico Cellinese (University of Florida), respectively. The proposal text is available online: Cellinese, Nico; Lapp, Hilmar (2015): An Ontology-Based System for Querying Life in a Post-Taxonomic Age. figshare. https://dx.doi.org/10.6084/m9.figshare.1401984
We are also very grateful to Ken and Linda McGurn for a gift in support of the project to Nico Cellinese.